This is a small exploration and best practices for reading data from disk.
One thing is for certain: base R reading and writing to disk are not optimal. However if your data is smallish (~ 5000 rows) the speeds don’t really matter that much. If you only need to load data once, convenience often triumphs speed.
If speed begins to matter, you have multiple options. In base there are
options like read.csv load readRDS. Or you could use another
package. There is data.table, readr, fst or the crosslanguage package
feather. Alternatively there are 2 embedded databases that you could
use, although reading an entire table sort of defeats the purpose of a
database.
In this overview I will show the read performance of
- utils::read.csv
- base::readRDS
- base::read.csv
- base::load
- data.table::fread
- feather::read_feather
- readr::read_csv
- readr::readRDS (wrapper around readRDS but with uncompressed file)
- fst::read.fst
- reading a table from SQLite
- reading a table from MonetDBlite
Apart from this benchmark of an entire dataset, there are other considerations for efficiency in your work. If you only need a few columns or rows from a dataset the databases and fst are excellent choices: they enable you to read in only a selection of columns and rows, without reading in the complete file first. If you do aggregations with window functions the two databases are excellent choices.
Preperation
First we create the dataset
set.seed(23455)
testdataset <-
data.frame(replicate(10, sample(0:2000, 15 * 10^5, rep = TRUE)),
replicate(10, stringi::stri_rand_strings(1000, 5)))
colnames(testdataset) <- tolower(gsub(pattern = "\\.",replacement = "_",colnames(testdataset)))# monetdb doesn't accept special characters or upper case without special treamtent.
and set the datapaths
# fs is a better worked out package to work with files.
if(!fs::dir_exists("data")){fs::dir_create("data")}
path_csv <- 'data/df.csv'
path_feather <- 'data/df.feather'
path_rdata <- 'data/df.RData'
path_rds <- 'data/df.rds'
path_rds_uncompressed <- 'data/df_uncompressed.rds'
path_fst <- 'data/df.fst'
path_sqlite <- "data/df.sqlite"
path_monetdblite <- "data/monetdb"
and load libraries.
# packages for reading and writing
library(feather)
library(data.table)
library(fst)
library(RSQLite)
library(MonetDBLite)
library(readr)
# packages for file manipulations plotting and benchmark
library(fs)
library(DBI)
library(microbenchmark)
library(ggplot2)
and write the files away
# set up database first
consqlite <- DBI::dbConnect(RSQLite::SQLite(),path_sqlite)
conmonet <- DBI::dbConnect(MonetDBLite::MonetDBLite(),path_monetdblite)
# write the files
DBI::dbWriteTable(consqlite,"sqlitedump",testdataset, overwrite = TRUE)
DBI::dbWriteTable(conmonet, "monetdbdump",testdataset, overwrite = TRUE)
#write.csv(testdataset, file = path_csv, row.names = F) # this is too slow, so
write_csv(testdataset, path_csv)
write_feather(testdataset, path_feather)
save(testdataset, file = path_rdata)
saveRDS(testdataset, path_rds)
readr::write_rds(testdataset, path_rds_uncompressed)
write.fst(testdataset, path_fst)
File sizes of saved files
While disk space is usually cheap, sometimes sizes matter. The saveRDS function uses compression by default. FST has a percentage compression you can set and is by default 50.
files <- list.files("data",full.names = TRUE)
info <- file.info(files)
info$size_mb <- info$size/(1024 * 1024)
print(subset(info, select=c("size_mb")),digits = 2)
## size_mb
## data/df_uncompressed.rds 114.56589
## data/df.csv 149.42059
## data/df.feather 114.52881
## data/df.fst 34.34178
## data/df.RData 28.54956
## data/df.rds 28.54952
## data/df.sqlite 139.87109
## data/monetdb 0.00015
# sum(fs::dir_info("data/monetdb/")$size) 1.72 kb
Benchmark reading times
Reading the same file 10 times. Do realise that for the databases this is the worst possible usecase: the database is meant to give you back a subset or calculate and give you the results.
suppressMessages({ # readr is too chatty for this overview
benchmark <- microbenchmark(
readCSV = utils::read.csv(path_csv),
readrCSV = readr::read_csv(path_csv, progress = F),
fread = data.table::fread(path_csv, showProgress = F),
loadRdata = base::load(path_rdata),
readRds = base::readRDS(path_rds),
readRDSuncompressed = readr::read_rds(path_rds_uncompressed),
readFeather = feather::read_feather(path_feather),
readFST = fst::read.fst(path_fst),
readSQLite = DBI::dbReadTable(consqlite, "sqlitedump"),
readMONETlite = DBI::dbReadTable(conmonet, "monetdbdump"),
times = 10)
})
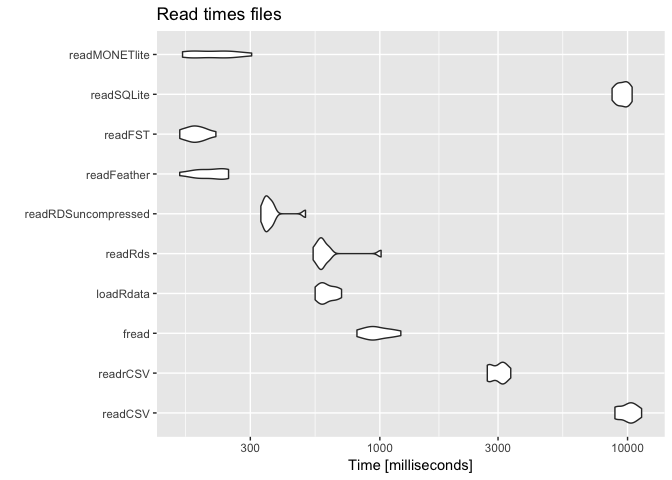
print(benchmark, signif = 2)
## Unit: milliseconds
## expr min lq mean median uq max neval
## readCSV 8900 9200 10000 10000 11000 11000 10
## readrCSV 2700 2800 3000 3100 3200 3400 10
## fread 810 910 980 960 1100 1200 10
## loadRdata 550 580 610 600 660 700 10
## readRds 540 570 620 580 610 1000 10
## readRDSuncompressed 330 340 370 350 370 500 10
## readFeather 160 180 210 210 240 250 10
## readFST 160 170 180 180 200 220 10
## readSQLite 8700 9000 9600 9600 10000 10000 10
## readMONETlite 160 170 220 210 250 300 10
Visualize results:
Recommondations for reading files
The recommodations for reading files, on this moment 2019-03-13 , are thus: If you are only using R, don’t write and read CSV. If you must use CSV and you are working with data.table use fread.
FST, Feather, readRDS (use without compression), loadRdata are all useful.
When to use a database?
If you take slices of data a database is almost always the better choice. SQLite and Monetdblite (probably others as well, but I really don’t want to start a db war here) are great choices for a data backend on disk. Monetdb has limitations on the column names, but is extremely fast in aggregations and according to many benchmarks (they really love benchmarking) monetdblite is much faster than sqlite. It all depends on what you want to do with the data. If you have a shiny app where you want to show subsets of data generate queries from the app to an embedded database.
Benchmark for writing files
I only did 5 repetitions for this one, because, boy, this is slow!
suppressMessages({ # readr is too chatty for this overview
benchmark_write <- microbenchmark(
#writeCSV = utils::write.csv(testdataset,path_csv),
writeCSV = readr::write_csv(testdataset,path_csv),
fwrite = data.table::fwrite(testdataset, path_csv),
saveRdata = base::save(testdataset,file = path_rdata),
#saveRds = base::saveRDS(testdataset, path_rds),
saveRDSuncompressed = readr::write_rds(testdataset, path_rds_uncompressed),
writeFeather = feather::write_feather(testdataset,path_feather),
writeFST = fst::write.fst(testdataset, path_fst),
writeSQLite = DBI::dbWriteTable(consqlite,"sqlitedump",testdataset, overwrite = TRUE),
writeMONETlite = DBI::dbWriteTable(conmonet, "monetdbdump",testdataset, overwrite = TRUE),
times = 5)
})
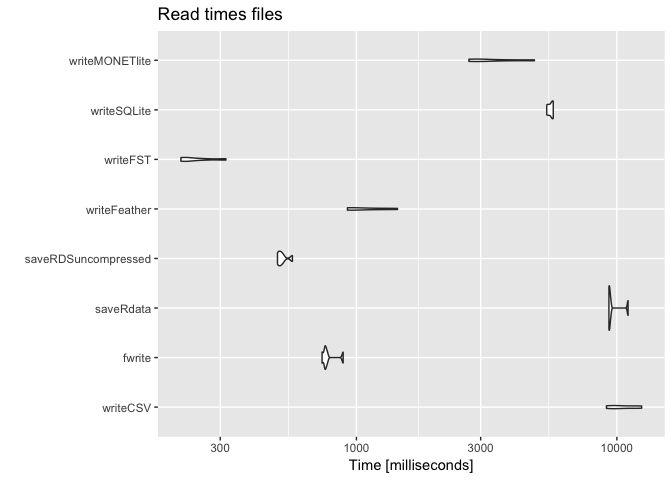
print(benchmark_write, signif = 2)
## Unit: milliseconds
## expr min lq mean median uq max neval
## writeCSV 9100 9700 11000 10000 12000 12000 5
## fwrite 740 760 780 760 770 890 5
## saveRdata 9300 9300 9700 9400 9500 11000 5
## saveRDSuncompressed 500 500 520 510 520 570 5
## writeFeather 920 930 1100 970 1200 1400!5
## writeFST 210 220 250 220 260 320 5
## writeSQLite 5400 5500 5600 5700 5700 5700 5
## writeMONETlite 2700 2900 3400 3000 3700 4800 5
Note that for databases there is often a highly optimized csv reader
Visualize results:
Recommondations for writing files
MonetDBLite::monetdblite_shutdown()
DBI::dbDisconnect(consqlite)
fs::dir_delete("data")
Versions are important here. My machine was like this:
sessioninfo::session_info()
## ─ Session info ──────────────────────────────────────────────────────────
## setting value
## version R version 3.5.1 (2018-07-02)
## os macOS 10.14.3
## system x86_64, darwin18.2.0
## ui X11
## language (EN)
## collate en_US.UTF-8
## ctype en_US.UTF-8
## tz Europe/Amsterdam
## date 2019-03-12
##
## ─ Packages ──────────────────────────────────────────────────────────────
## package * version date lib
## assertthat 0.2.0 2017-04-11 [2]
## bit 1.1-14 2018-05-29 [1]
## bit64 0.9-7 2017-05-08 [1]
## blob 1.1.1 2018-03-25 [1]
## cli 1.0.1 2018-09-25 [2]
## codetools 0.2-16 2018-12-24 [3]
## colorspace 1.4-0 2019-01-13 [1]
## crayon 1.3.4 2017-09-16 [2]
## data.table * 1.12.1 2019-03-08 [1]
## DBI * 1.0.0.9001 2019-01-29 [1]
## digest 0.6.18 2018-10-10 [2]
## dplyr 0.8.0.1 2019-02-15 [1]
## evaluate 0.13 2019-02-12 [1]
## feather * 0.3.2 2019-01-07 [1]
## fs * 1.2.6 2018-08-23 [2]
## fst * 0.8.10 2018-12-14 [1]
## ggplot2 * 3.1.0 2018-10-25 [2]
## glue 1.3.0 2018-07-17 [2]
## gtable 0.2.0 2016-02-26 [2]
## hms 0.4.2 2018-03-10 [2]
## htmltools 0.3.6 2017-04-28 [2]
## knitr 1.22 2019-03-08 [1]
## lazyeval 0.2.1 2017-10-29 [2]
## magrittr 1.5 2014-11-22 [2]
## memoise 1.1.0 2017-04-21 [2]
## microbenchmark * 1.4-6 2018-10-18 [1]
## MonetDBLite * 0.6.0 2018-07-27 [1]
## munsell 0.5.0 2018-06-12 [2]
## pillar 1.3.1 2018-12-15 [1]
## pkgconfig 2.0.2 2018-08-16 [2]
## plyr 1.8.4 2016-06-08 [2]
## purrr 0.3.1 2019-03-03 [1]
## R6 2.4.0 2019-02-14 [2]
## Rcpp 1.0.0 2018-11-07 [2]
## readr * 1.3.1 2018-12-21 [1]
## rlang 0.3.1 2019-01-08 [1]
## rmarkdown 1.11 2018-12-08 [2]
## RSQLite * 2.1.1.9002 2019-01-29 [1]
## scales 1.0.0 2018-08-09 [2]
## sessioninfo 1.1.1 2018-11-05 [2]
## stringi 1.3.1 2019-02-13 [2]
## stringr 1.4.0 2019-02-10 [1]
## tibble 2.0.1 2019-01-12 [1]
## tidyselect 0.2.5 2018-10-11 [2]
## withr 2.1.2 2018-03-15 [2]
## xfun 0.5 2019-02-20 [1]
## yaml 2.2.0 2018-07-25 [2]
## source
## CRAN (R 3.5.1)
## CRAN (R 3.5.1)
## CRAN (R 3.5.1)
## CRAN (R 3.5.1)
## CRAN (R 3.5.1)
## CRAN (R 3.5.1)
## CRAN (R 3.5.1)
## CRAN (R 3.5.1)
## local
## Github (r-dbi/DBI@c7333be)
## CRAN (R 3.5.1)
## CRAN (R 3.5.1)
## CRAN (R 3.5.1)
## CRAN (R 3.5.1)
## CRAN (R 3.5.1)
## CRAN (R 3.5.1)
## CRAN (R 3.5.1)
## CRAN (R 3.5.1)
## CRAN (R 3.5.1)
## CRAN (R 3.5.1)
## CRAN (R 3.5.1)
## CRAN (R 3.5.1)
## CRAN (R 3.5.1)
## CRAN (R 3.5.1)
## CRAN (R 3.5.1)
## CRAN (R 3.5.1)
## CRAN (R 3.5.1)
## CRAN (R 3.5.1)
## CRAN (R 3.5.1)
## CRAN (R 3.5.1)
## CRAN (R 3.5.1)
## CRAN (R 3.5.1)
## CRAN (R 3.5.1)
## CRAN (R 3.5.1)
## CRAN (R 3.5.1)
## CRAN (R 3.5.1)
## CRAN (R 3.5.1)
## Github (rstats-db/RSQLite@c347308)
## CRAN (R 3.5.1)
## CRAN (R 3.5.1)
## CRAN (R 3.5.1)
## CRAN (R 3.5.1)
## CRAN (R 3.5.1)
## CRAN (R 3.5.1)
## CRAN (R 3.5.1)
## CRAN (R 3.5.1)
## CRAN (R 3.5.1)
##
## [1] /Users/roelhogervorst/Library/R/3.x/library
## [2] /usr/local/lib/R/3.5/site-library
## [3] /usr/local/Cellar/r/3.5.1/lib/R/library
Further reading
- https://appsilon.com/fast-data-loading-from-files-to-r/ “From 2017-04-11, used the same data and approach for reading files, but not all of these options”
- https://kbroman.org/blog/2017/04/30/sqlite-feather-and-fst/ “2017-04-30, another comparison between fst, sqlite and feather, also looks into write performance”